Plate Zip and Index file
The plate zip and index file are the inputs needed to run KREAP.
Plate ZIP
Create a folder with every well you want to be included in your analysis:
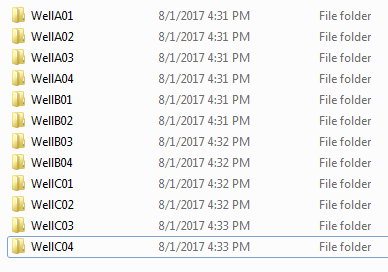
Create a new zip of these folders:
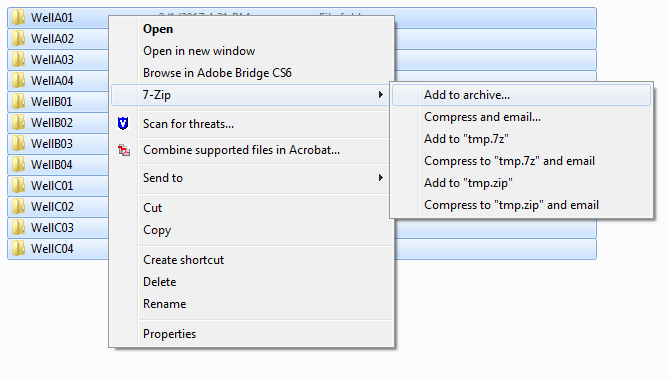
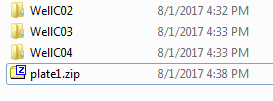
The newly created zip is one of the two inputs for KREAP:
Index
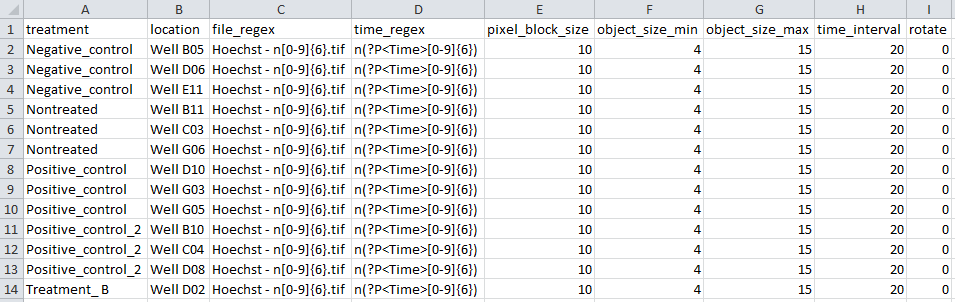
The index file has the following structure:
| Column Name | Description |
|---|---|
| treatment | The treatment group |
| location | Location of the well (relative in the zip) |
| file_regex | A regular expression that helps ignore other (irrelevant) files in a well |
| time_regex | A regular expression that selects the time point of an image |
| pixel_block_size | The pixel block size parameter for Cellprofiler |
| object_size_min | The minimum object size (in pixels) |
| object_size_max | The maximum object size (in pixels) |
| time_interval | The time (in minutes) between images |
| rotate | The images will be rotated (clockwise) by this many degrees (optional) |
Here is the index file associated with the zip created in the previous section:
The index file doesn’t have to be created in Excel, but it’s convenient.
Any other program that can output tabular files should also work (including any regular text editors).
Note
It’s possible to create zip and index file that contain multiple plates by using extra directories in the zip and index file:
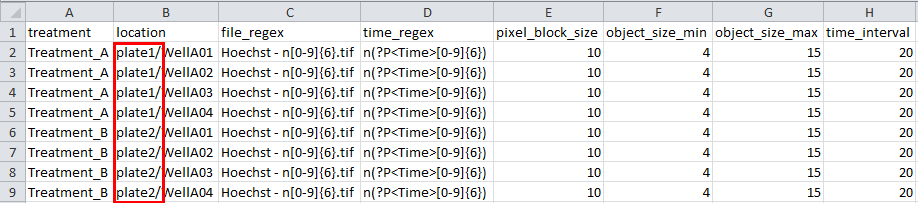
Example
We have prepared a plate zip and index files that can directly be used with KREAP or can be used as a template for your own data:
Sample Zip
Hoechst Plate Index
CellTracker Plate Index
Analyse your data with KREAP
Uploading file to Galaxy KREAP
Run KREAP Image Analysis
Run KREAP Data Modeling
Handling errors in the modeling result